[4]:
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
from jupyterthemes import jtplot
plt.style.use('mike')
jtplot.style(context='talk', fscale=1, grid=False)
import cosmogrb
GRBs¶
This section describes how to handle the low level simulation of GRBs. AS the code currently is built for simulating GRBs as observed by Fermi-GBM, we will focus our attention there. As the code expands, I will update the docs.

Instantiate the GRB with its parameters¶
For this example, we will create a GRB that has its flux coming from a single pulse shape that is described by a cutoff power law evolving in time.
[2]:
grb = cosmogrb.gbm.GBMGRB_CPL(
ra=312.0,
dec=-62.0,
z=1.0,
peak_flux=5e-7,
alpha=-0.66,
ep=500.0,
tau=2.0,
trise=1.0,
tdecay=1.0,
duration=80.0,
T0=0.1,
)
grb.info()
| 0 | |
|---|---|
| name | SynthGRB |
| z | 1 |
| ra | 312 |
| dec | -62 |
| duration | 80 |
| T0 | 0.1 |
| 0 | |
|---|---|
| peak_flux | 5.000000e-07 |
| alpha | -6.600000e-01 |
| trise | 1.000000e+00 |
| tdecay | 1.000000e+00 |
| 0 |
|---|
Examine the latent lightcurve¶
[3]:
time = np.linspace(0, 20, 500)
grb.display_energy_integrated_light_curve(time, color="#A363DE");
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
<ipython-input-3-0737622692d6> in <module>
1 time = np.linspace(0, 20, 500)
2
----> 3 grb.display_energy_integrated_light_curve(time, color="#A363DE");
4
~/coding/projects/cosmogrb/cosmogrb/grb/grb.py in display_energy_integrated_light_curve(self, time, ax, **kwargs)
143 """
144
--> 145 list(self._lightcurves.values())[0].display_energy_integrated_light_curve(
146 time=time, ax=ax, **kwargs
147 )
IndexError: list index out of range
[ ]:
energy = np.logspace(1, 3, 1000)
grb.display_energy_dependent_light_curve(time, energy, cmap='PRGn', lw=.25, alpha=.5)
Simulate the GRB¶
Now we can create all the light curves from the GRB. Since are not currently running a Dask server, we tell the GRB to process serially, i.e., computing each light curve one at a time.
[ ]:
grb.go(serial=True)
Save the GRB to an HDF5 file¶
As this is a time-consuming operation, we want to be able to save the GRB to disk. This is done by serializing all the light curves and information about the GRB into an HDF5 file.
[ ]:
grb.save('test_grb.h5')
Reload the GRB¶
What if want to reload the GRB? We need to create and instance of GRBSave from the file we just created. Notice all the information about the GRB is recovered.
[5]:
grb_reload = cosmogrb.GRBSave.from_file('test_grb.h5')
grb_reload.info()
| 0 | |
|---|---|
| name | SynthGRB |
| z | 1 |
| ra | 312 |
| dec | -62 |
| duration | 80 |
| T0 | 0.1 |
| 0 | |
|---|---|
| alpha | -6.600000e-01 |
| peak_flux | 5.000000e-07 |
| tdecay | 1.000000e+00 |
| trise | 1.000000e+00 |
The GRBSave contents¶
The stores all the information about the light curves and the instrument responses used to generate the data. Each light curve/ response pair can be accessed as keys of the GRBSave. Then one can easily, examine/plot/process the contents of each light curve.
[6]:
for key in grb_reload.keys():
lightcurve = grb_reload[key]['lightcurve']
lightcurve.info()
| 0 | |
|---|---|
| name | b0 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76246e+08 |
| T0 | 0.1 |
| n_counts | 190656 |
| n_counts_source | 36 |
| n_counts_background | 190660 |
| 0 | |
|---|---|
| angle | 106.678116 |
| 0 | |
|---|---|
| name | b1 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76219e+08 |
| T0 | 0.1 |
| n_counts | 206738 |
| n_counts_source | 70 |
| n_counts_background | 206695 |
| 0 | |
|---|---|
| angle | 64.599896 |
| 0 | |
|---|---|
| name | n0 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76253e+08 |
| T0 | 0.1 |
| n_counts | 194080 |
| n_counts_source | 136 |
| n_counts_background | 194008 |
| 0 | |
|---|---|
| angle | 140.423466 |
| 0 | |
|---|---|
| name | n1 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76276e+08 |
| T0 | 0.1 |
| n_counts | 198594 |
| n_counts_source | 127 |
| n_counts_background | 198530 |
| 0 | |
|---|---|
| angle | 123.192564 |
| 0 | |
|---|---|
| name | n2 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76203e+08 |
| T0 | 0.1 |
| n_counts | 203221 |
| n_counts_source | 87 |
| n_counts_background | 203189 |
| 0 | |
|---|---|
| angle | 87.337115 |
| 0 | |
|---|---|
| name | n3 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76206e+08 |
| T0 | 0.1 |
| n_counts | 200535 |
| n_counts_source | 108 |
| n_counts_background | 200460 |
| 0 | |
|---|---|
| angle | 120.135933 |
| 0 | |
|---|---|
| name | n4 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76229e+08 |
| T0 | 0.1 |
| n_counts | 204014 |
| n_counts_source | 89 |
| n_counts_background | 203953 |
| 0 | |
|---|---|
| angle | 115.423396 |
| 0 | |
|---|---|
| name | n5 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76212e+08 |
| T0 | 0.1 |
| n_counts | 201485 |
| n_counts_source | 47 |
| n_counts_background | 201489 |
| 0 | |
|---|---|
| angle | 91.866668 |
| 0 | |
|---|---|
| name | n6 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76284e+08 |
| T0 | 0.1 |
| n_counts | 203790 |
| n_counts_source | 121 |
| n_counts_background | 203697 |
| 0 | |
|---|---|
| angle | 126.89814 |
| 0 | |
|---|---|
| name | n7 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76264e+08 |
| T0 | 0.1 |
| n_counts | 205853 |
| n_counts_source | 142 |
| n_counts_background | 205745 |
| 0 | |
|---|---|
| angle | 150.685788 |
| 0 | |
|---|---|
| name | n8 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76264e+08 |
| T0 | 0.1 |
| n_counts | 205156 |
| n_counts_source | 127 |
| n_counts_background | 205069 |
| 0 | |
|---|---|
| angle | 121.106339 |
| 0 | |
|---|---|
| name | n9 |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76252e+08 |
| T0 | 0.1 |
| n_counts | 198283 |
| n_counts_source | 85 |
| n_counts_background | 198226 |
| 0 | |
|---|---|
| angle | 108.143856 |
| 0 | |
|---|---|
| name | na |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76287e+08 |
| T0 | 0.1 |
| n_counts | 198458 |
| n_counts_source | 582 |
| n_counts_background | 197918 |
| 0 | |
|---|---|
| angle | 57.909913 |
| 0 | |
|---|---|
| name | nb |
| instrument | GBM |
| tstart | -100 |
| tstop | 300 |
| time adjustment | 5.76277e+08 |
| T0 | 0.1 |
| n_counts | 206218 |
| n_counts_source | 206 |
| n_counts_background | 206091 |
| 0 | |
|---|---|
| angle | 77.225671 |
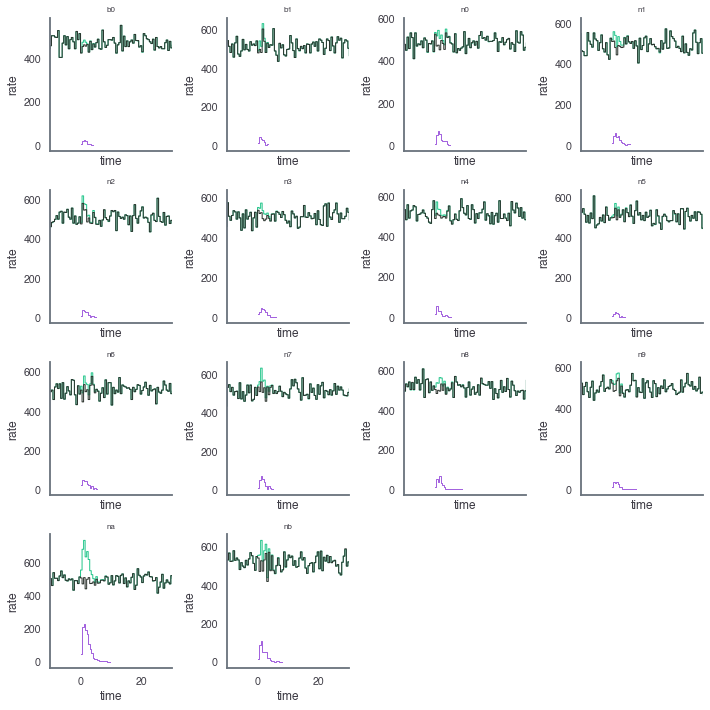
For example, let’s look at the total, source, and background data light curves generated.
[9]:
fig, axes = plt.subplots(4,4,sharex=True,sharey=False,figsize=(10,10))
row=0
col = 0
for k,v in grb_reload.items():
ax = axes[row][col]
lightcurve =v['lightcurve']
lightcurve.display_lightcurve(dt=.5, ax=ax,lw=1,color='#25C68C')
lightcurve.display_source(dt=.5,ax=ax,lw=1,color="#A363DE")
lightcurve.display_background(dt=.5,ax=ax,lw=1, color="#2C342E")
ax.set_xlim(-10, 30)
ax.set_title(k,size=8)
if col < 3:
col+=1
else:
row+=1
col=0
axes[3,2].set_visible(False)
axes[3,3].set_visible(False)
plt.tight_layout()
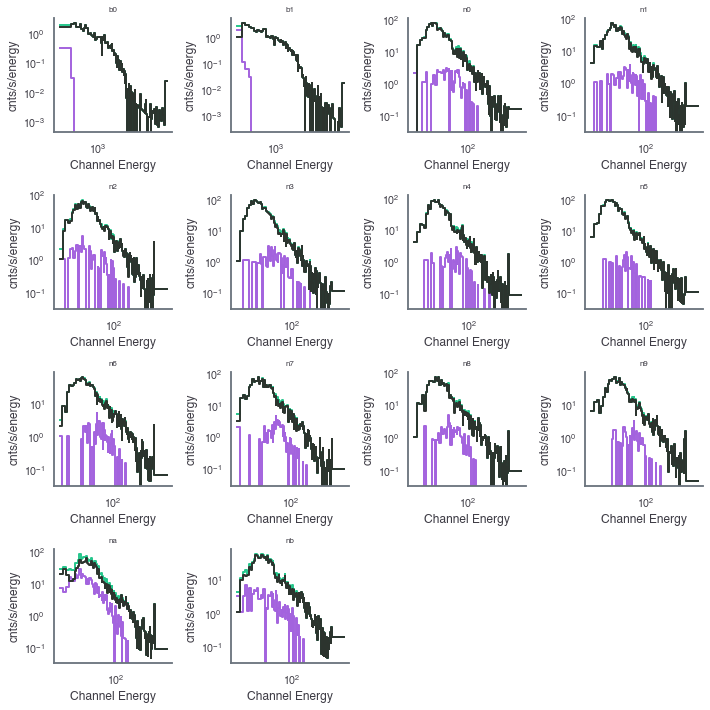
And we can look at the generated count spectra/
[10]:
fig, axes = plt.subplots(4,4,sharex=False,sharey=False,figsize=(10,10))
row=0
col = 0
for k, v in grb_reload.items():
ax = axes[row][col]
lightcurve = v['lightcurve']
lightcurve.display_count_spectrum(tmin=0, tmax=5, ax=ax,color='#25C68C')
lightcurve.display_count_spectrum_source(tmin=0, tmax=5, ax=ax,color="#A363DE")
lightcurve.display_count_spectrum_background(tmin=0, tmax=5, ax=ax, color="#2C342E")
ax.set_title(k,size=8)
if col < 3:
col+=1
else:
row+=1
col=0
axes[3,2].set_visible(False)
axes[3,3].set_visible(False)
plt.tight_layout()
Convert HDF5 to standard FITS files¶
In the case of GBM, we can convert the saved HDF5 files into TTE files for analysis in 3ML.
[12]:
cosmogrb.grbsave_to_gbm_fits("test_grb.h5")
!ls SynthGRB_*
ls: SynthGRB_*: No such file or directory
[ ]: